Home
iVenomDB provides the scientific community with the insect venom protein sequences and other functional information, mainly derived from literature curation. The large quantity of sequences and annotations are valuable resources for researchers to investigate aspects of venom gene evolution and applications The user-friendly interface and online tools will help interested researchers retrieve the information they want easily.
[1] Entries of ten parts of iVenomDB.
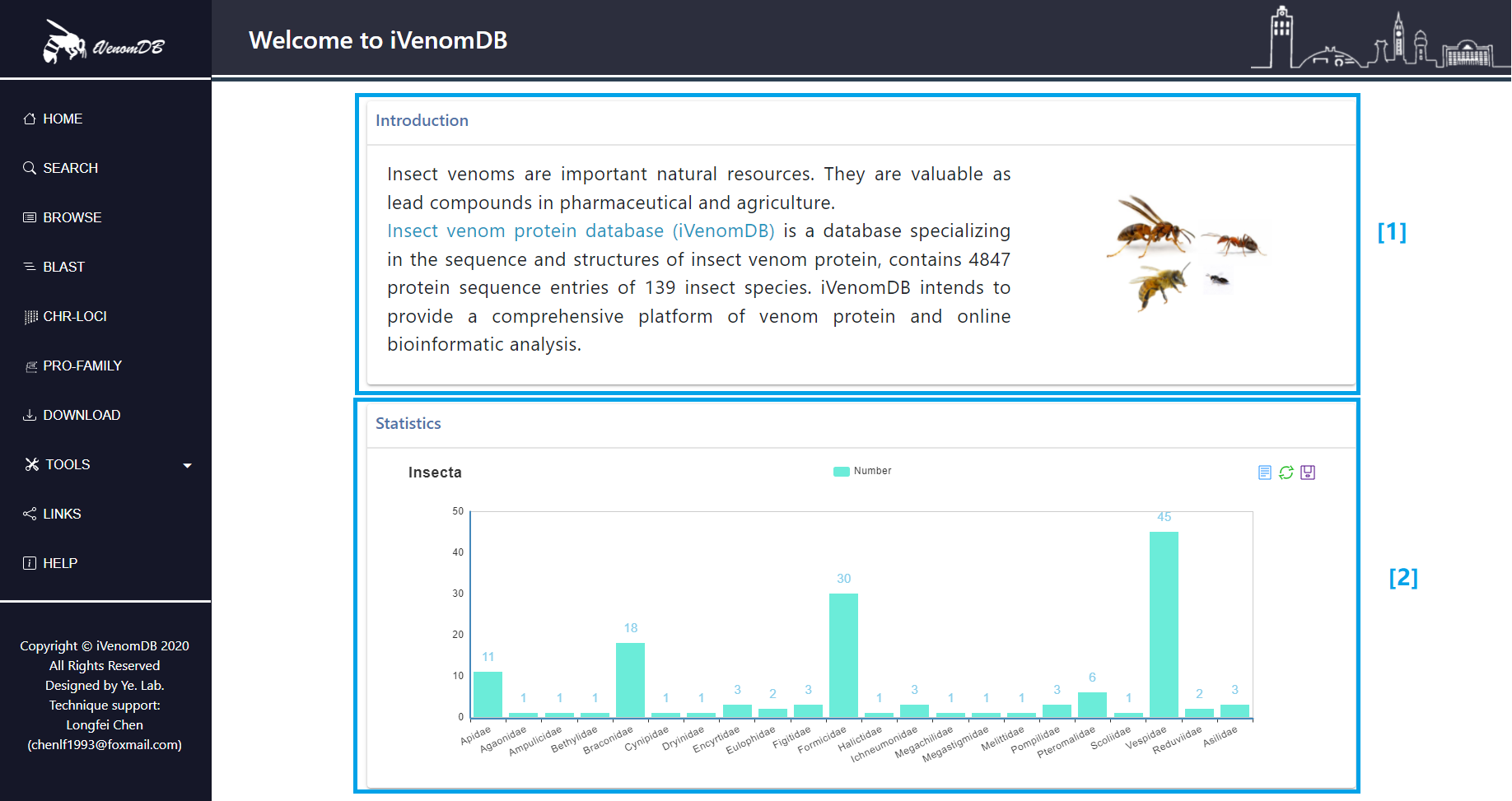
[2] The statistics of insect species of different families.
Search
In the page, species can be selected from the species list. Some frequently studied protein names are displayed by frequency ranks and can be clicked to enter into the search box automatically. The search results are listed in a table and can be clicked to link to venom protein entry card page.
Search Input
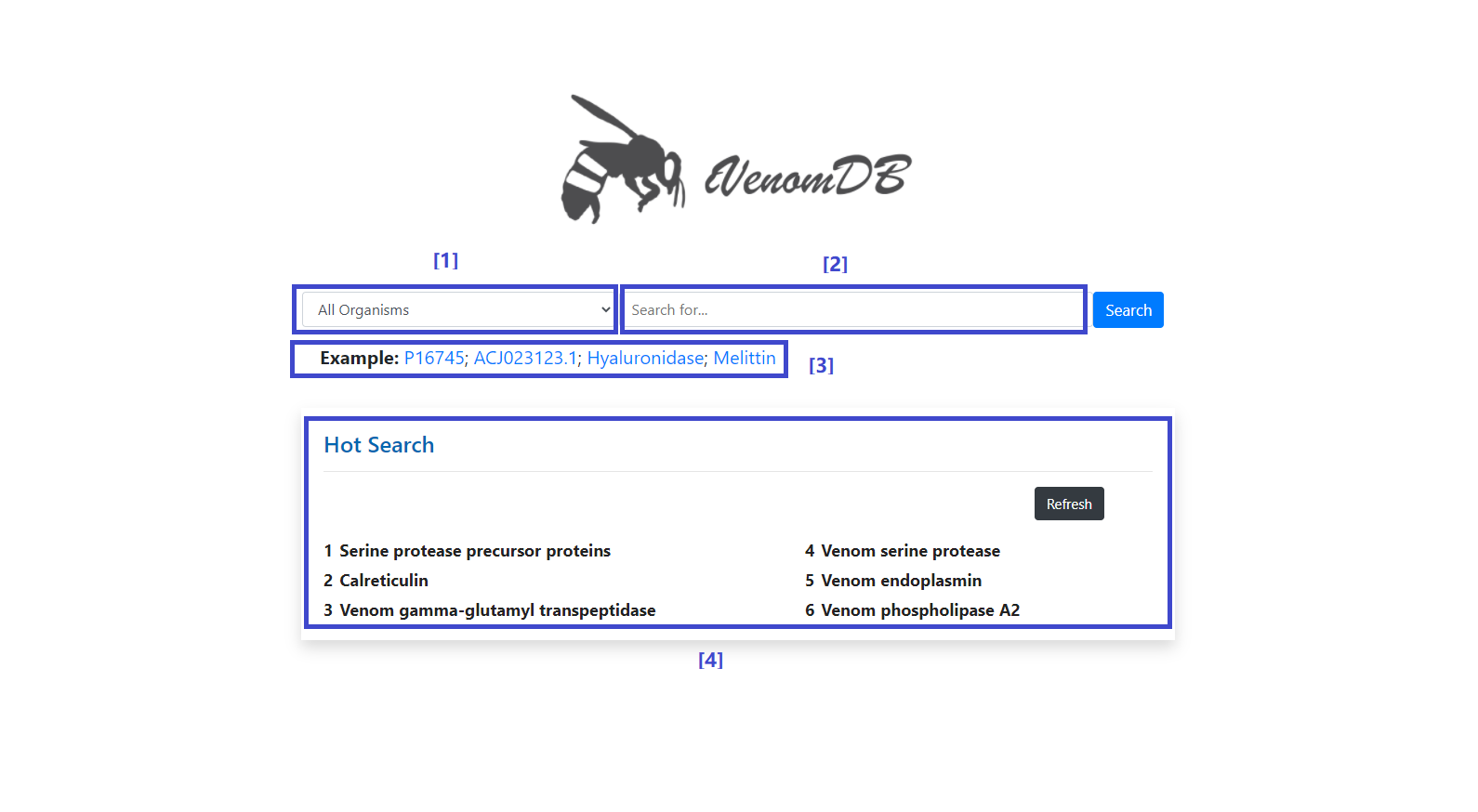
[1] Species list of insects in database.
[2] Enter the search terms.
[3] The frequent terms can be selected.
Search Output
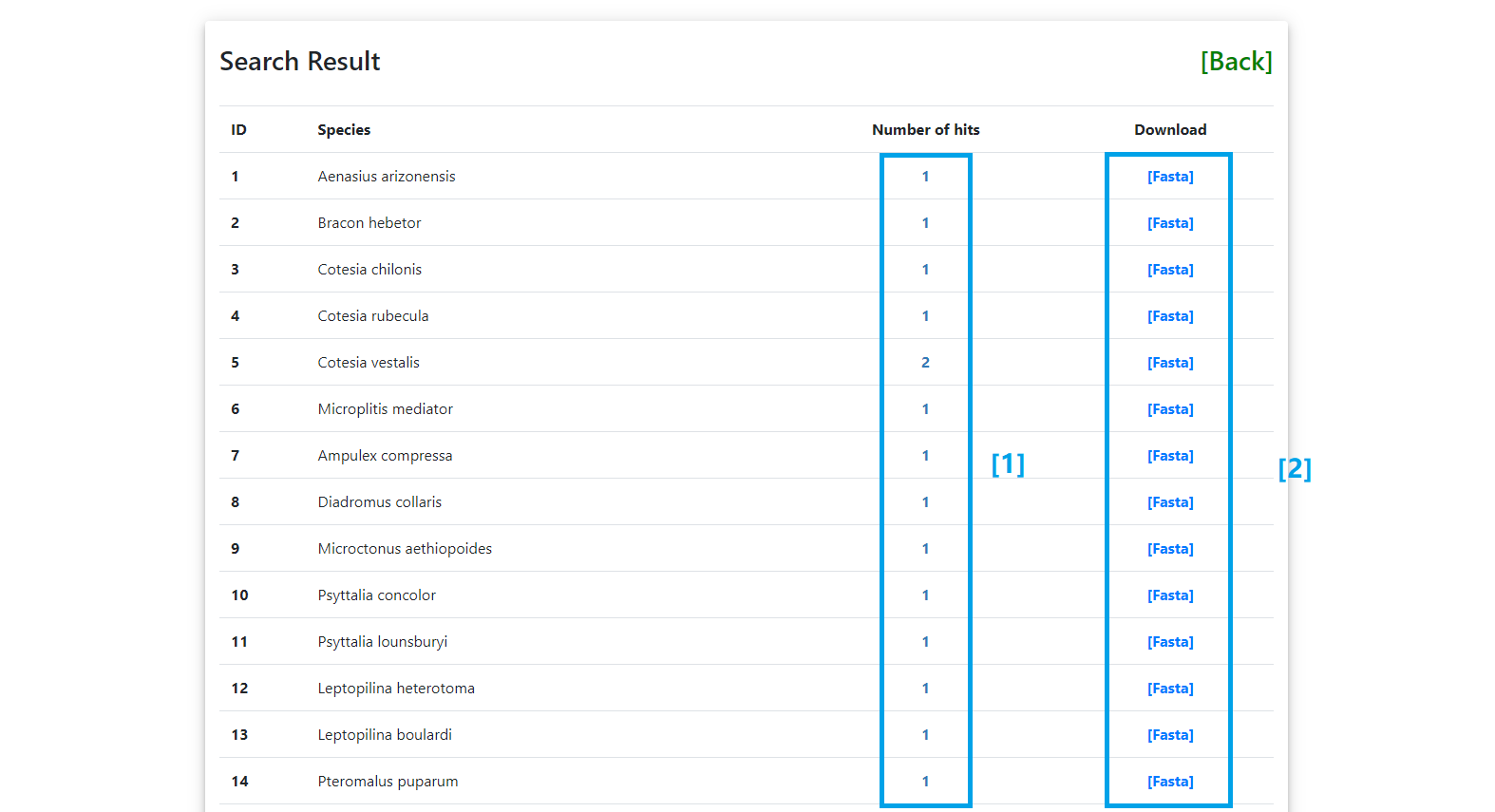
[1] Click here to show the details of search results of the selected species.
[2] Click here to download the sequences directly.
[1] Click here to link to venom protein card.
Browse
Venom proteins are sorted by species and displayed on a dynamic tree. The trees are constructed according to taxonomic relationships of species. After clicking the species name, information including species name, taxonomy, brief introduction, and photos will displayed on the right. A table will list all the venom proteins of this species below. It also includes entry IDs, full names, short names and lengths.
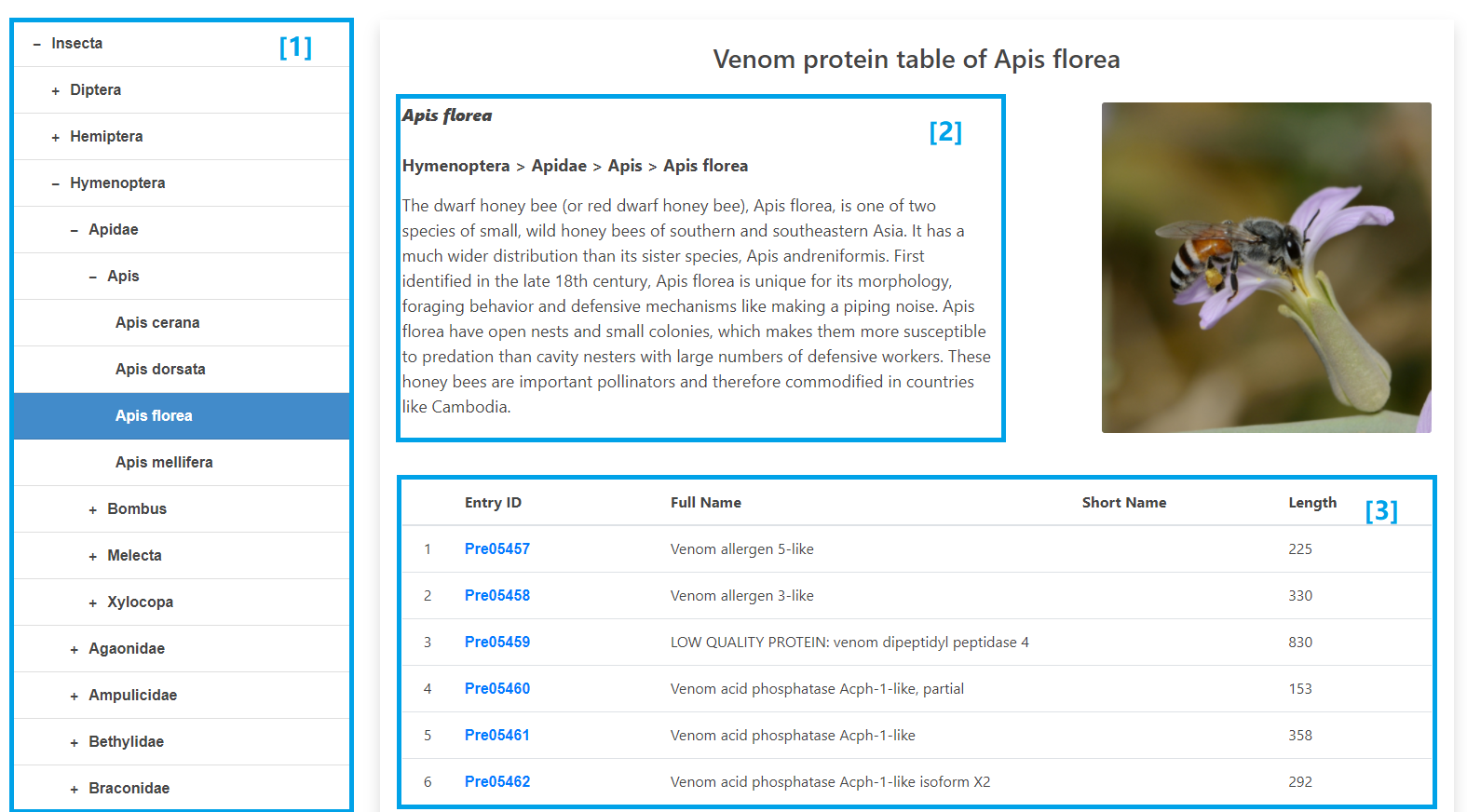
[1] Click “+” to expand and click “-” to collapse. Select species to show all its venom proteins.
[2] The basic information of selected species.
[3] Table lists all the venom proteins of this species. It includes entry IDs, full names, short names and lengths.
Venom Protein Card
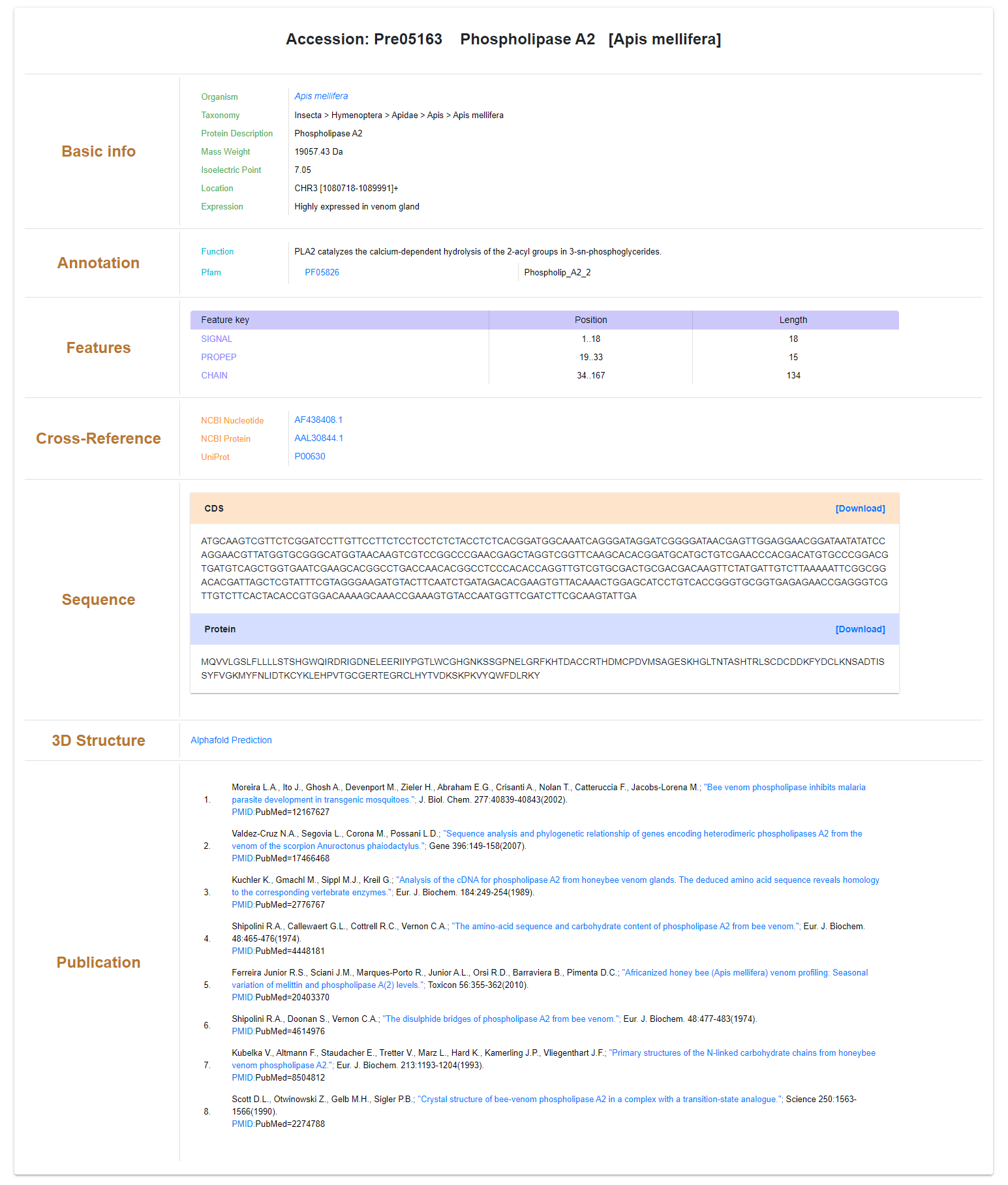
All information for each venom protein entry is displayed in property-value card format. The source species, its taxonomy, physical properties (pI and MW), genetic location and expression specificity are provided on the first part of the page. The annotation part displays the functions and other functional annotations of the venom protein. Position ranges of signal peptide, propeptide and mature protein sequence are displayed in a table. The positions and types of PTMs are also listed. The protein and its corresponding reference nucleotide sequences are downloadable from the sequence part in FASTA format. Cross-reference of accessions from NCBI, UniProtKB, PDB and other databases can be clicked to see details. Publications reporting the venom with all details are available to interested readers.
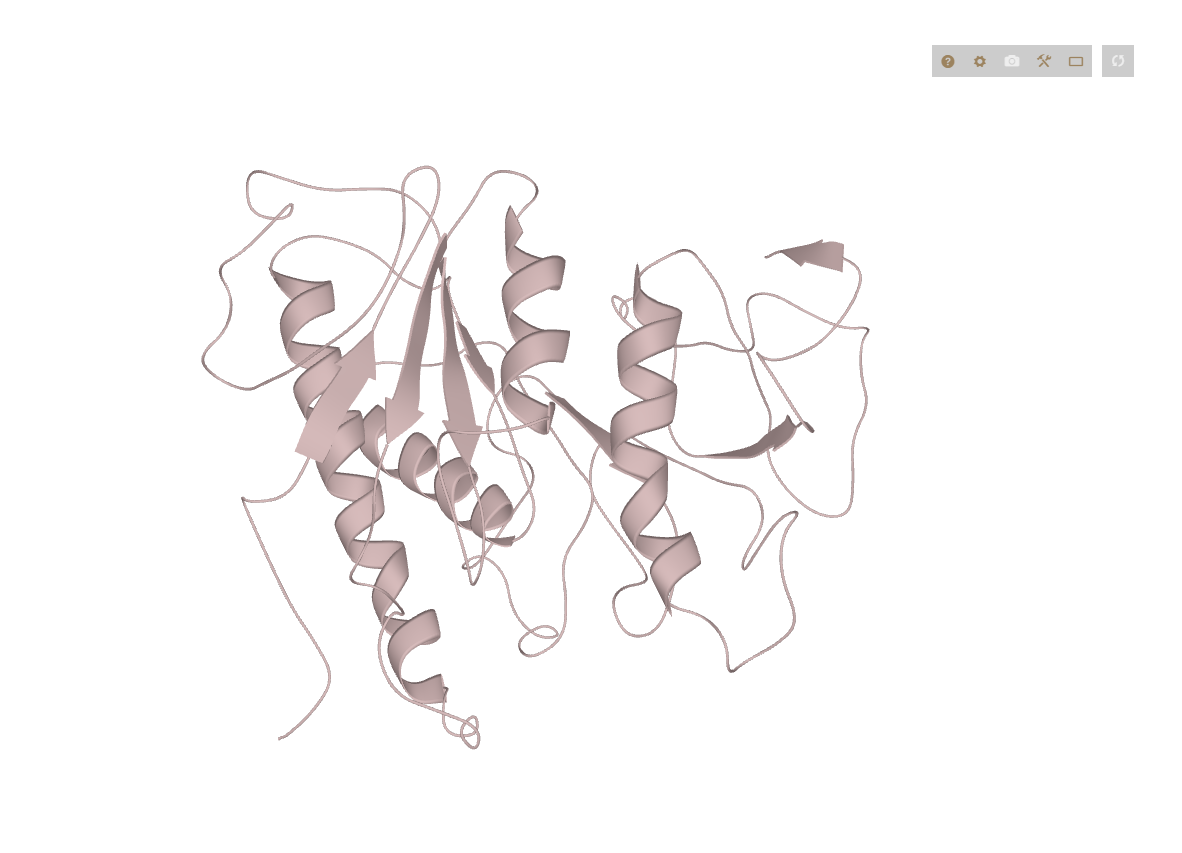
3D Structure
Protein 3D structures are displayed as interactive 3D models.
Blast
Sequence similarity is an important principle to retrieve proteins or genes when there is only partial sequence available for searching homologues. Diamond software, with higher speed, will run when it receives user commands. For searching the proteins, the software within blastp and blastx is enough to complete all the tasks. Similarly, the blast result is also displayed in a table with some scores for each row of result.
Blast Input
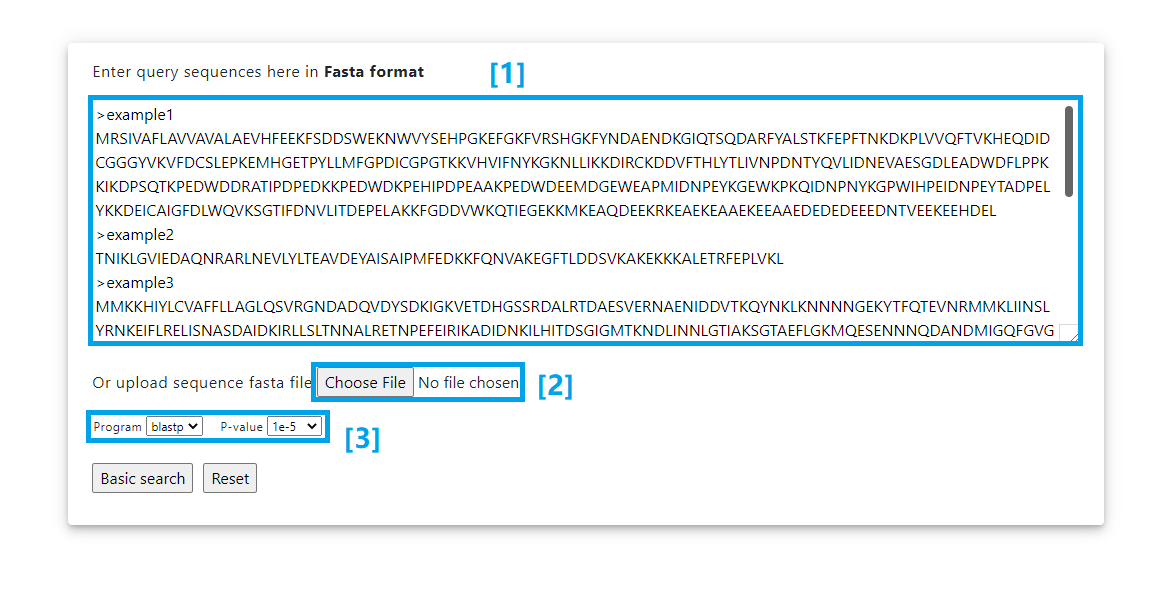
[1] Paste your fasta format sequences into the box.
[2] Or input the search sequence file here.
[3] Choose the blast program and the p value.
Blast Output
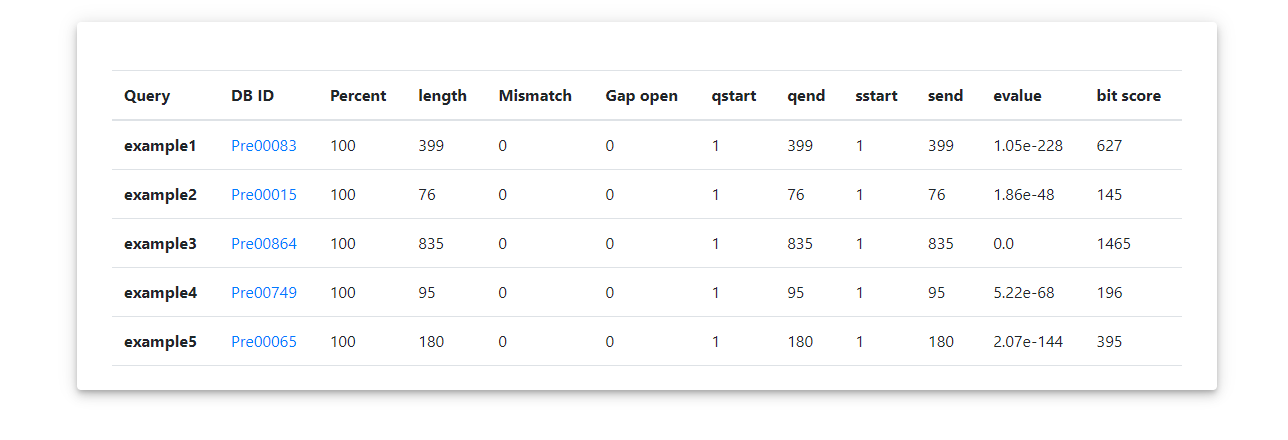
The page show the blast result list.
CHR-LOCI
Venom genes are generally recruited by gene duplication and other mechanisms. Therefore, the positions of venom genes on chromosomes are important for studying the venom gene evolution. For some venomous insects with chromosome-level genome assemblies, venom genes are displayed on the loci. Users can select the species of interest from the species list options. Hovering the mouse over the genes will display the detailed positions. Clicking the gene will link to the go to entry card page

[1] Choose the species list with chromosome level assembly with venom gene on it.
[2] Click the venom gene on chromosome to show the venom protein detail.
PRO-FAMILY
All representative protein families are visualized as donut charts. The different colors represent the four insect groups, wasps, ants, bees and hornets. The areas stand for the proportion of genes in the family. Clicking the specific protein family will pull up the phylogenetic tree.
PRO-FAMILY
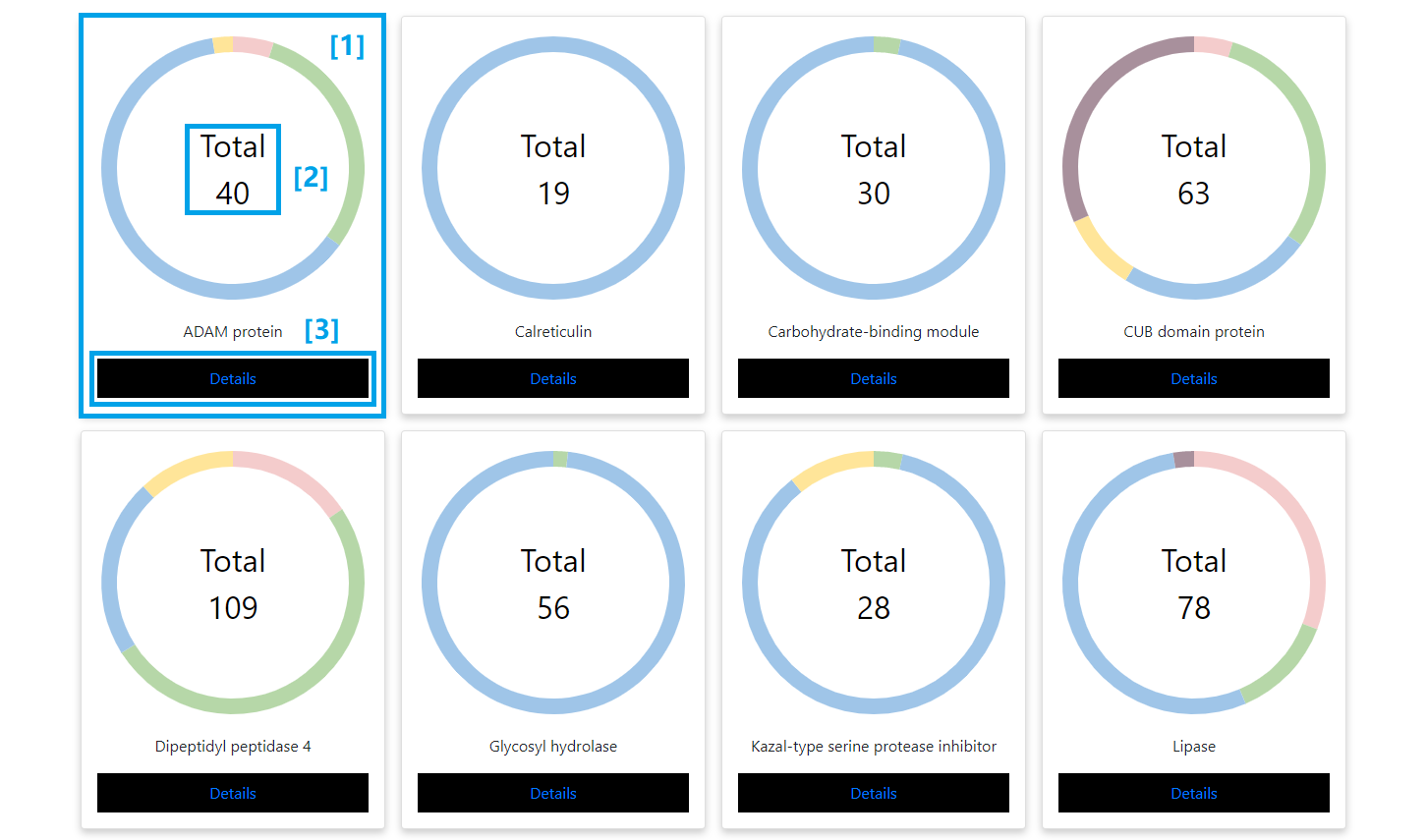
[1] Each donut chart represent a venom protein family.
[2] The number shows the total number of venom protein in the selected family.
[3] Click here to show the phylogenetic tree of the selected protein family.
Phylogenetic Tree
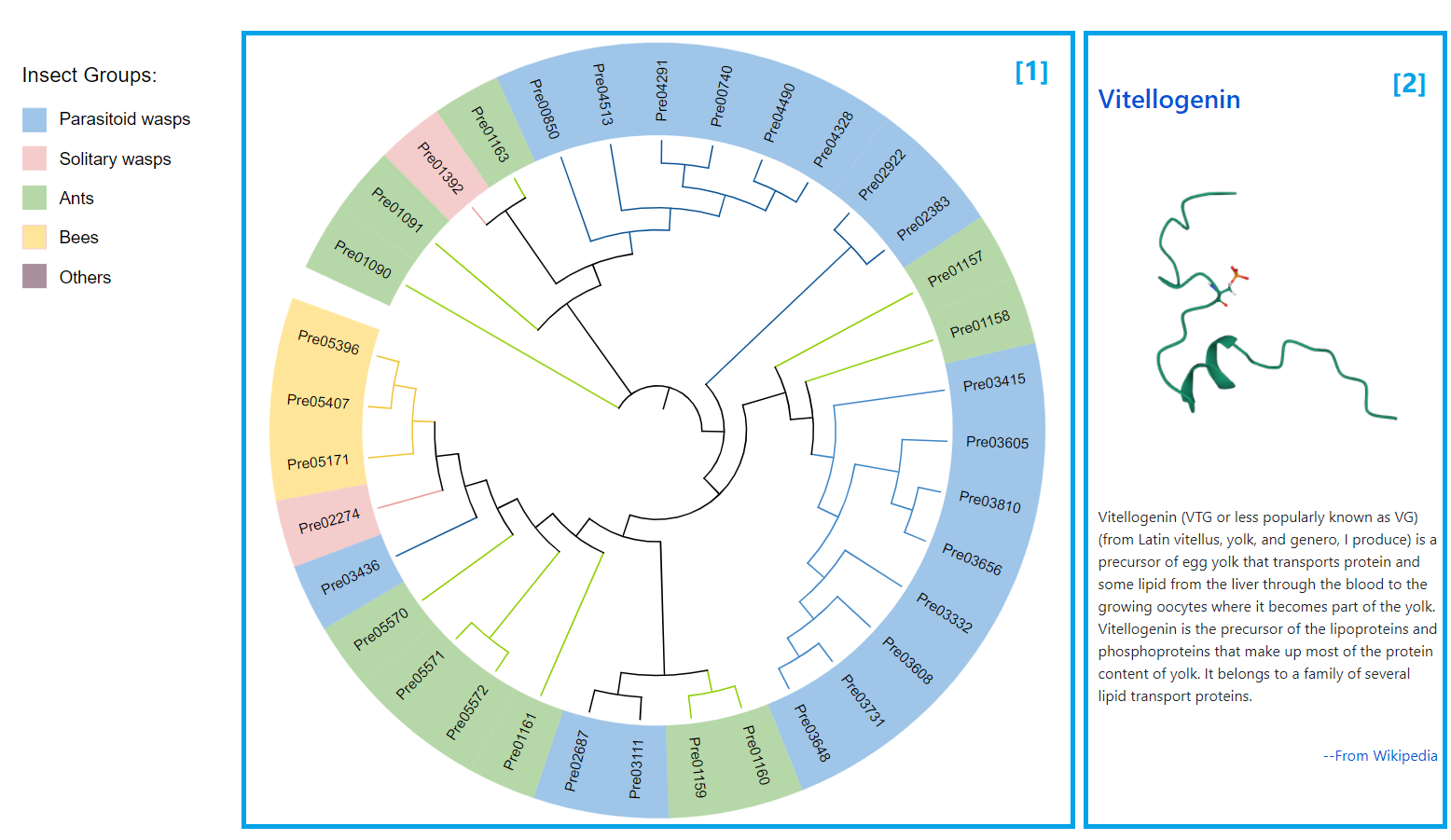
[1] The phylogenetic tree of the selected family. The venom genes are displayed on the tree and can be clicked to link to the entry card page.
[2] The basic information of the selected family.
Download
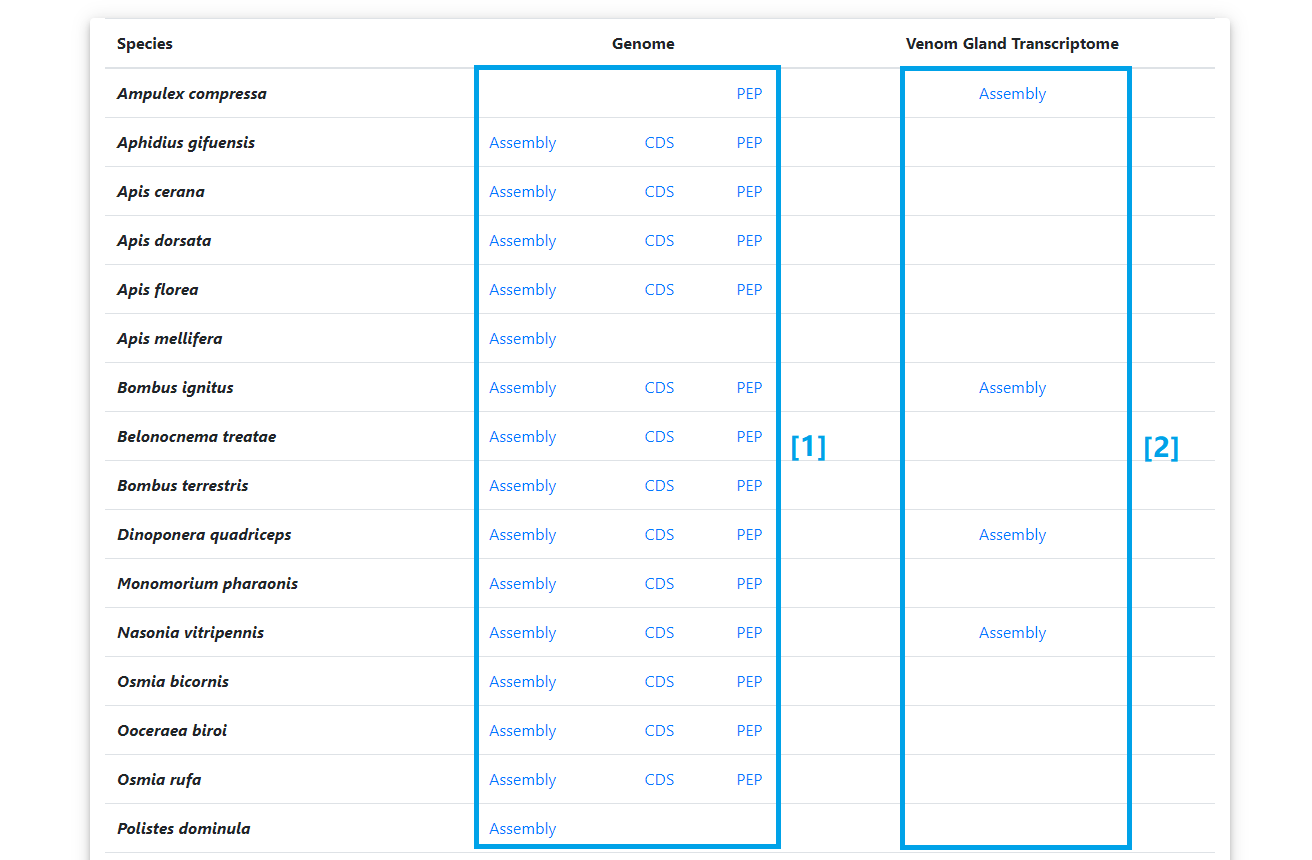
[1] The genome assembly, official gene set and protein sequences of select species.
[2] The transcript assembly of venom gland transcriptome.
[3] The mass spectrometry detected peptide sequences in venom proteome.
Tools
Tools are provided for users to carry out custom analysis online. This part includes the application of tools, such as mafft, trimal and signalp.
Links

The page list all the related databases and introductions.
